Notebooks
Premium
Trends
BioTuring
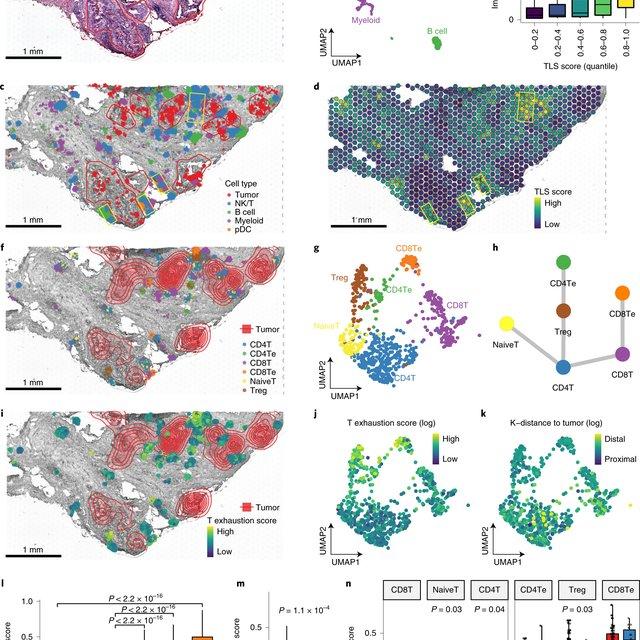
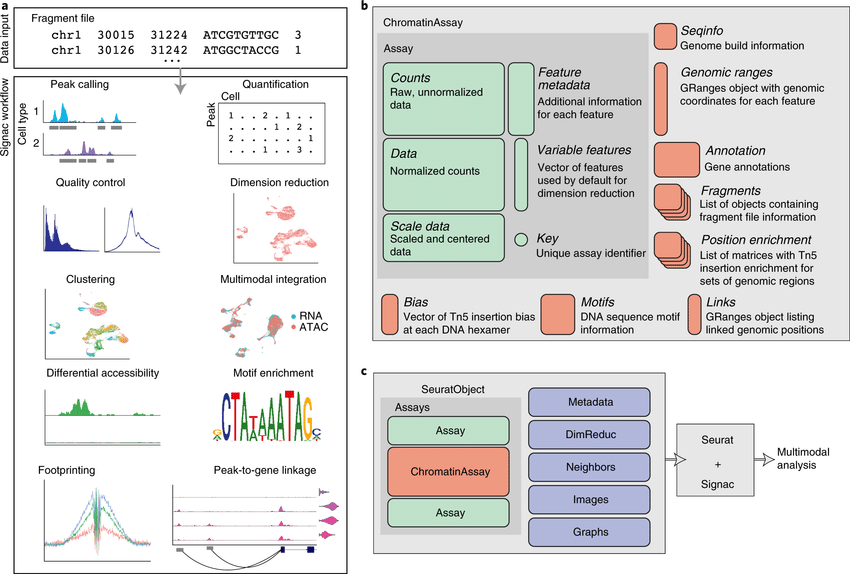
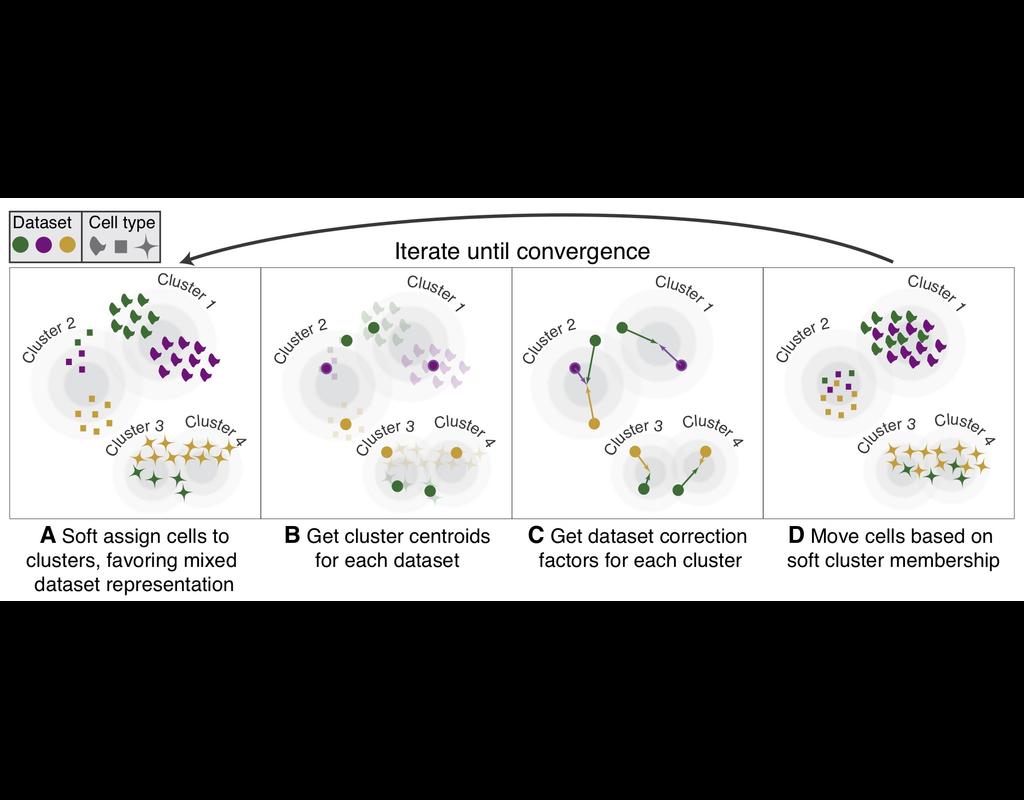
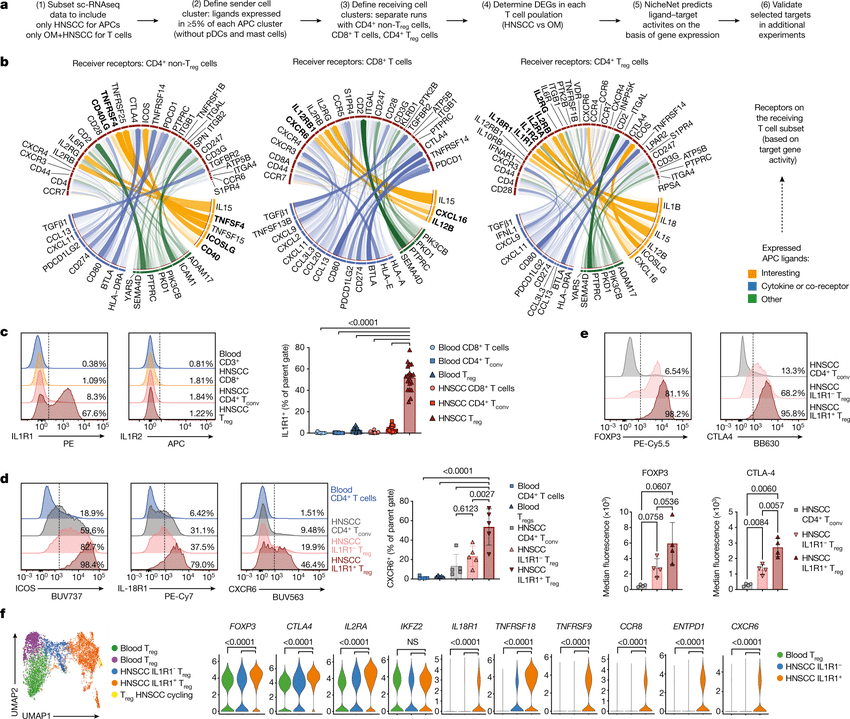
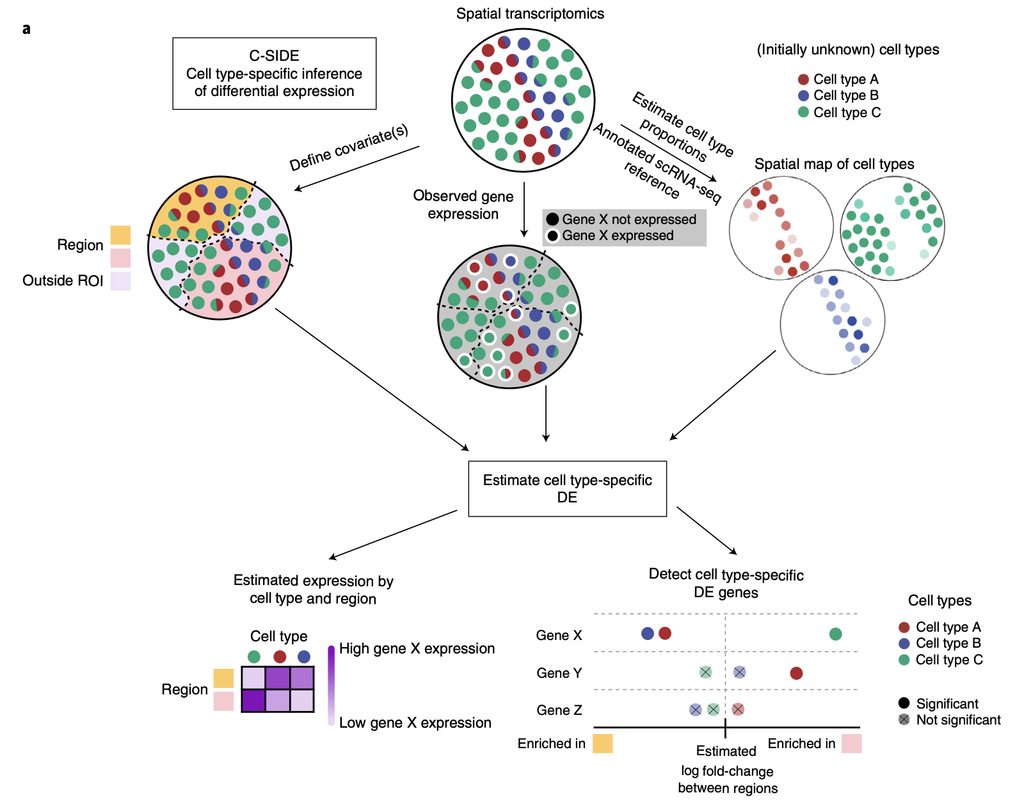
Recent spatial transcriptomics (ST) technologies have allowed us to capture cellular heterogeneity while retaining spatial information. However, ST datasets may lose single-cell resolution, limiting the discovery of cell-type-specific spatial pattern(More)