Notebooks
Premium
Trends
BioTuring
BioTuring
BioTuring
BioTuring
BioTuring
BioTuring
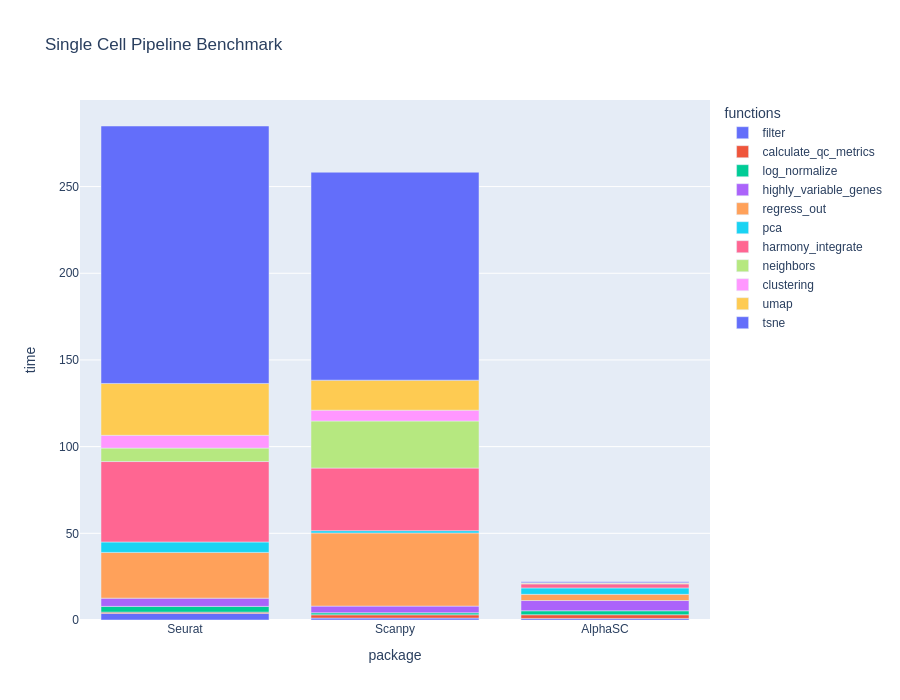
BioTuring
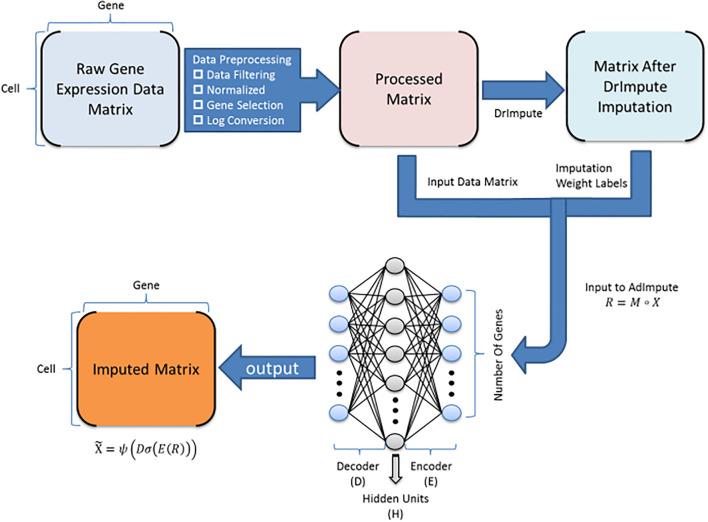
BioTuring
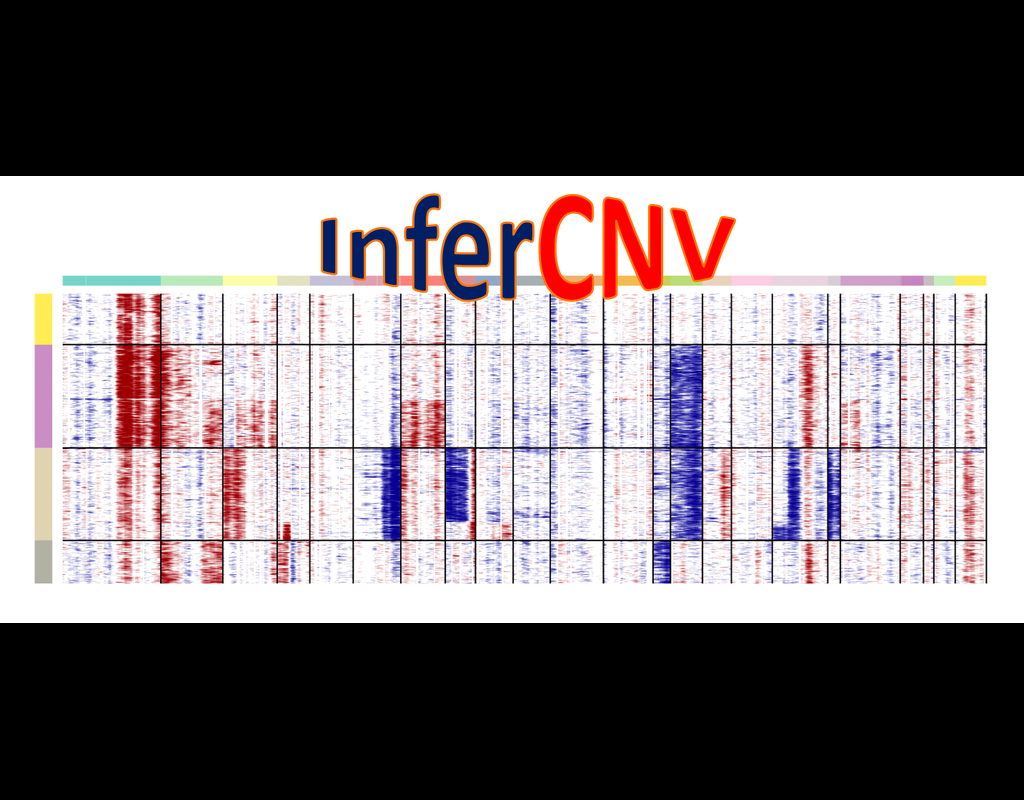
BioTuring
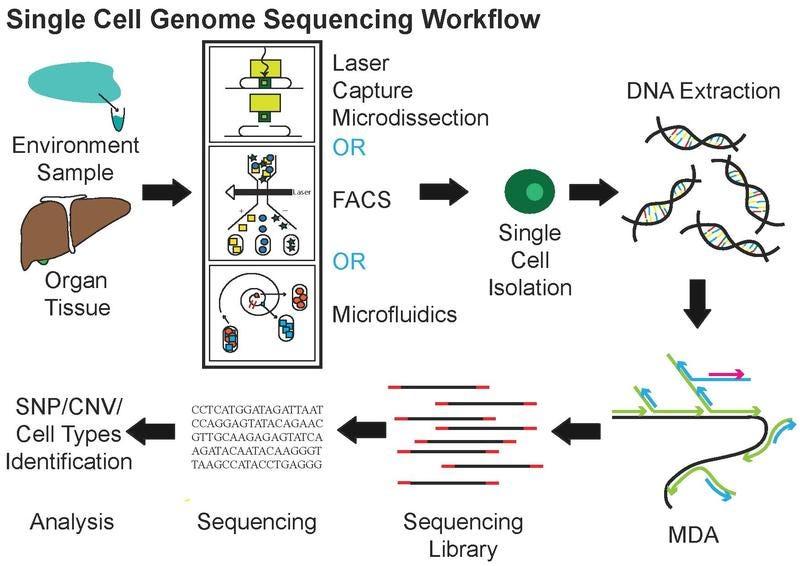
BioTuring
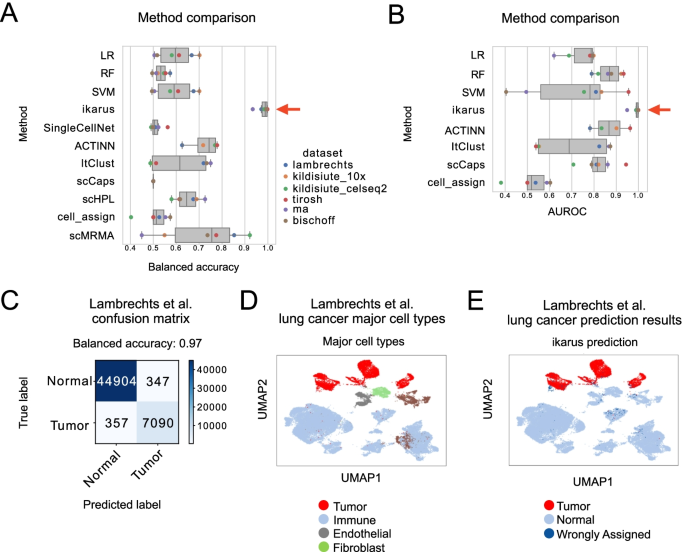
BioTuring
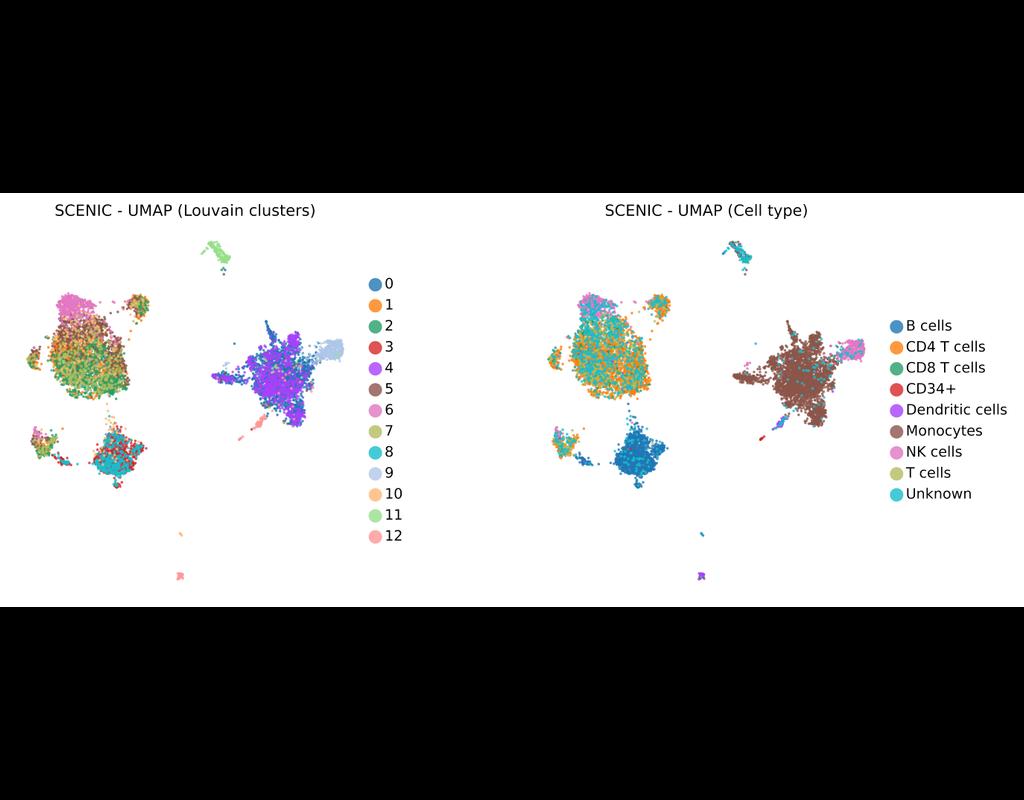
BioTuring
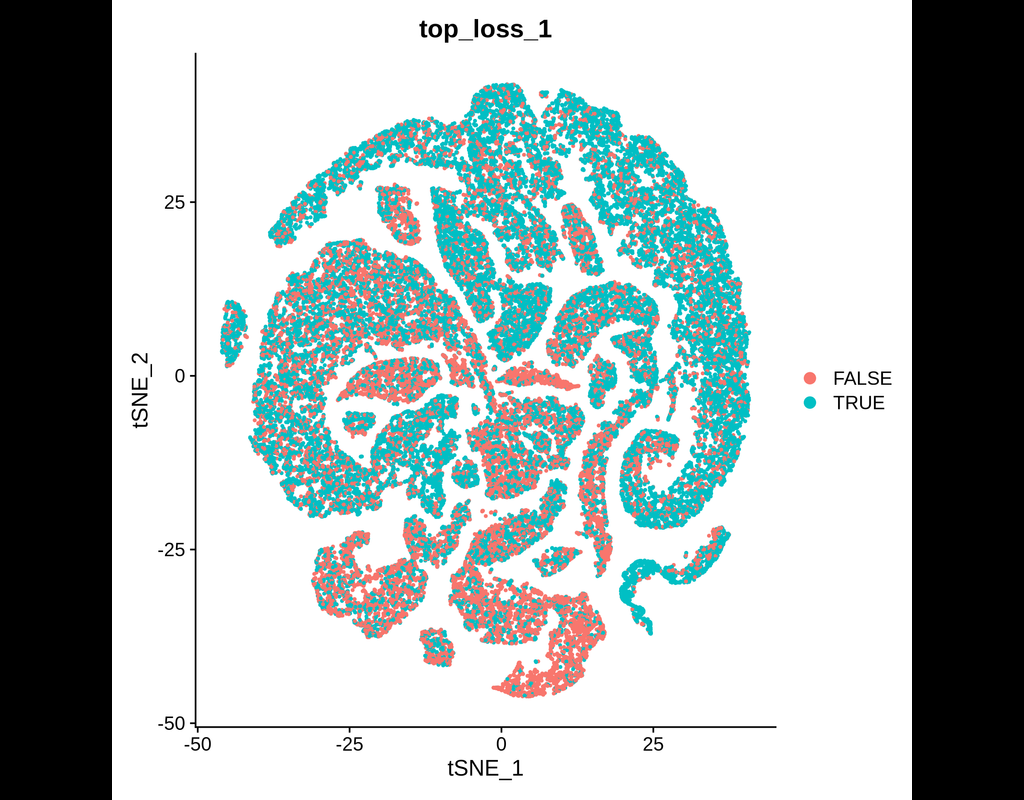
BioTuring
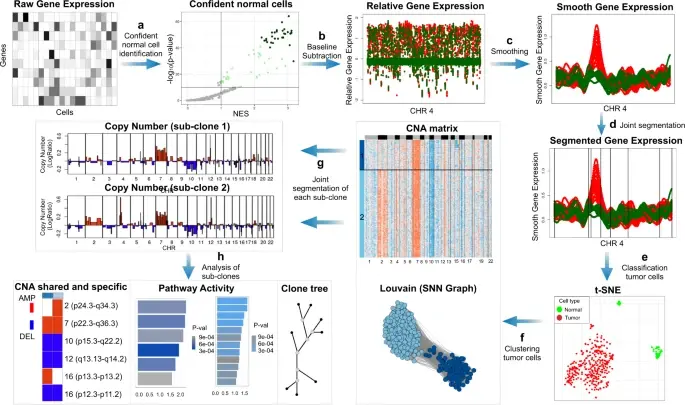
BioTuring
BioTuring
| Notebooks |
|---|